Large Language Models (LLMs) are well-suited for working with non-structured data. So far, their usage with structured data hasn’t been explored in depth although structured data is everywhere relational databases are. Making an LLM able to interact with a relational database can be an interesting idea since it will unlock the possibility of letting a user “chat with the data” and let the LLM discover relationships inside the data lake
In this article, we’ll explore a possible integration of Gemini (the multimodal large language model developed by Google) with PostgreSQL, and how to build a Retrieval-Augmented Generation (RAG) system to navigate in the structured data. Everything will be done using the Go programming language.
This is the fourth article of a series about the usage of Vertex AI in Go, and as such, it will share the same prerequisite presented in both of them: the services account creation, the environment variables, and so on. The prerequisite parts can be read in each of those articles.
- Custom model training & deployment on Google Cloud using Vertex AI in Go
- AutoML pipeline for tabular data on VertexAI in Go.
- Using Gemini in a Go application: limits and details
This article tries to implement the idea presented at the end of the last article. Given that converting all the user data to a textual representation overflows the maximum context length of Gemini, we implement a RAG to overcome this limitation.
RAG and Embeddings
Before going to the implementation in PostgreSQL, Go, and Gemini (through Vertex AI) we need to understand how a RAG system works. The analogy with a detective searching inside a massive document archive for clues works well. In a RAG we have three components:
- The Detective: This is the generative model, like Gemini, that uses its knowledge to answer your questions or complete tasks.
- The Archive: This is your PostgreSQL database, holding all the tabular data (your documents).
- The Informant: This is the retriever, a special tool that understands both your questions and the data in the archive. It acts like your informant, scanning the archive to find the most relevant documents (clues) for the detective.
But how does the informant know which documents are relevant? Here’s where embeddings come in. Embeddings are like condensed summaries of information. Imagine each document and your questions are shrunk down into unique sets of numbers. The closer these numbers are in space, the more similar the meaning.
The informant uses an embedding technique to compare your question’s embedding to all the document embeddings in the archive. It then retrieves the documents with the most similar embeddings, essentially pointing the detective in the right direction.
With these relevant documents at hand, the detective (generative model) can then analyze them and use its knowledge to answer your question or complete your request.
Given this structure, we need:
- The detective: in our case, it will be Gemini used through Vertex AI.
- The embedding model: a model able to create embeddings from a document.
- The archive: PostgreSQL. We need to convert the structured information from the database to a format valid for the embedding model. Then store the embeddings on the database.
- The informant: pgvector. The open-source vector similarity search extension for PostgreSQL.
The embedding model is able to create embeddings only of a document. So, we need to find a way to convert the structured representation into a document as the first step.
From structured to Unstructured data
LLMs are very good at extracting information from textual data and executing tasks described using text. Depending on our data, we may be lucky to have something easy “to narrate”.
In the case described in this article, we are going to use all the data related to sleep, physical activities, food, heart rate, and number of steps (and others) gathered during a day for a single user. With this information it’s quite easy to extract a regular description of a user’s day, section by section. Being the data so regular, we can try to make it fit in a template.
The template: the daily report
We can define a template that summarizes/highlights the important part we want to be able to retrieve while searching through our RAG. The template will be used by Gemini as part of its prompt in a chat session. In this chat session, we are going to ask the model to extract from the JSON data the information that we want to display in the report.
### Date [LLM to write date]
## Activity
- Total Active Time: [LLM to fill from activities_summaries.active_minutes]
- Calories Burned: [LLM to fill from activities_summaries.calories_out]
- Steps Taken: [LLM to fill from activities_summaries.steps]
- Distance Traveled: [LLM to fill from activities_summaries.distance / activities_summary_distances.distance]
- List of activities: [LLM to iterate through activities_summary_activities and fill name, duration, calories burned]
### Active Minutes Breakdown
- Lightly Active Minutes: [LLM to fill from activities_summaries.lightly_active_minutes]
- Fairly Active Minutes: [LLM to fill from activities_summaries.fairly_active_minutes]
- Very Active Minutes: [LLM to fill from activities_summaries.very_active_minutes]
### Heart Rate Zones
- [LLM to iterate through activities_summary_heart_rate_zones and fill from heart_rate_zones (zone name, minutes)]
## Sleep
- Total Sleep Duration: [LLM to fill from sleep_logs.duration]
- Sleep Quality: [LLM to fill from sleep_logs.efficiency]
- Deep Sleep: [LLM to fill from sleep_stage_details where sleep_stage='deep sleep'] (minutes)
- Light Sleep: [LLM to fill from sleep_stage_details where sleep_stage='light sleep'] (minutes)
- REM Sleep: [LLM to fill from sleep_stage_details where sleep_stage='rem sleep'] (minutes)
- Time to Fall Asleep: [LLM to fill from sleep_logs.minutes_to_fall_asleep]
## Exercise Activities
- [LLM to iterate through daily_activity_summary_activities / minimal_activities and fill name, duration, calories burned (from activity_logs)]
...
Before digging inside the Go code, we have to design the structure for our data in the database.
The simplest solution is to create a table containing the textual reports that our LLM will generate together with its “compact representation” (the embeddings).
The table creation
Being our data already stored on PostgreSQL it would be ideal to use the same database also for storing the embeddings and performing spatial queries on them, and not introduce a new “vector database”.
pgvector is the extension for PostgreSQL that allows us to define a data type “vector” and it gives our operators and function to perform measures like cosine distance, l2 distance, and many others.
Once installed and granted the superuser access to our database user, we can enable the extension and define the table for storing our data.
-- Enable the pgvector extension
CREATE EXTENSION IF NOT EXISTS vector;
CREATE TABLE IF NOT EXISTS reports (
id SERIAL PRIMARY KEY,
user_id BIGINT NOT NULL REFERENCES users(id),
start_date DATE NOT NULL,
end_date DATE NOT NULL,
report_type TEXT NOT NULL,
report TEXT NOT NULL,
embedding VECTOR
);
After enabling the vector extension we can define the embedding field of type vector. There’s no need to specify the maximum length of the vector since the extension supports dynamically shaped vectors.
The table is defined to store all the users’ reports. In this article, we are going to cover only the daily reports (so start_date will be equal to end_date) but to concept is easily generalizable to different kinds of reports. This is also the reason for the report_type field.
The Go data structure
It’s a good practice to map a SQL table to a struct. Using galeone/igor for interacting with PostgreSQL from Go this is pretty much mandatory.
import (
"time"
"github.com/pgvector/pgvector-go"
)
type Report struct {
ID int64 `igor:"primary_key"`
UserID int64
StartDate time.Time
EndDate time.Time
ReportType string
Report string
Embedding pgvector.Vector
}
func (r *Report) TableName() string {
return "reports"
}
That’s all. We are now ready to interact with Vertex AI to:
- Go from structured to unstructured data, making Gemini filling the previously defined template.
- Generate the embeddings of both the report.
- Let the user create a chat session with Gemini and create the embeddings of its prompt.
- Doing a spatial query for retrieving the (hopefully) relevant documents we have in the database.
- Pass these documents to Gemini as its search context.
- Ask the model to answer the user question by looking at the provided document.
The Reporter type
We can design a data type Reporter whose goal is to generate these reports. Using the (well-known after three articles) pattern for the interaction with Vertex AI, we are going to create 2 different clients:
- The generative AI client for Gemini
- The prediction client for our embedding model
import (
vai "cloud.google.com/go/aiplatform/apiv1beta1"
vaipb "cloud.google.com/go/aiplatform/apiv1beta1/aiplatformpb"
"cloud.google.com/go/vertexai/genai"
"google.golang.org/api/option"
"google.golang.org/protobuf/types/known/structpb"
)
type Reporter struct {
user *types.User
predictionClient *vai.PredictionClient
genaiClient *genai.Client
ctx context.Context
}
// NewReporter creates a new Reporter
func NewReporter(user *types.User) (*Reporter, error) {
ctx := context.Background()
var predictionClient *vai.PredictionClient
var err error
if predictionClient, err = vai.NewPredictionClient(ctx, option.WithEndpoint(_vaiEndpoint)); err != nil {
return nil, err
}
var genaiClient *genai.Client
const region = "us-central1"
if genaiClient, err = genai.NewClient(ctx, _vaiProjectID, region, option.WithCredentialsFile(_vaiServiceAccountKey)); err != nil {
return nil, err
}
return &Reporter{user: user, predictionClient: predictionClient, genaiClient: genaiClient, ctx: ctx}, nil
}
// Close closes the client
func (r *Reporter) Close() {
r.predictionClient.Close()
r.genaiClient.Close()
}
Our Reporter will be used to generate both the reports and its vector representation (embedding).
Generate the embeddings
We can start by using the predictionClient to invoke a text embedding model.
The pattern is always the same. Working with Vertex AI in Go is quite convoluted because every client request has to be created by filling in the correct protobuf fields and this is verbose and not immediate. Just look at all the boilerplate code we have to write to extract the embeddings from the response.
_vaiEmbeddingsEndpoint is the global variable containing the endpoint for the chosen model. In our case the endpoint for Google’s model textembedding-gecko@003.
This method returns a pgvector.Vector offered by the pgvector/pgvector-go package.
import "github.com/pgvector/pgvector-go"
// GenerateEmbeddings uses VertexAI to generate embeddings for a given prompt
func (r *Reporter) GenerateEmbeddings(prompt string) (embeddings pgvector.Vector, err error) {
var promptValue *structpb.Value
if promptValue, err = structpb.NewValue(map[string]interface{}{"content": prompt}); err != nil {
return
}
// PredictRequest: create the model prediction request
// autoTruncate: false
var autoTruncate *structpb.Value
if autoTruncate, err = structpb.NewValue(map[string]interface{}{"autoTruncate": false}); err != nil {
return
}
req := &vaipb.PredictRequest{
Endpoint: _vaiEmbeddingsEndpoint,
Instances: []*structpb.Value{promptValue},
Parameters: autoTruncate,
}
// PredictResponse: receive the response from the model
var resp *vaipb.PredictResponse
if resp, err = r.predictionClient.Predict(r.ctx, req); err != nil {
return
}
// Extract the embeddings from the response
mapResponse, ok := resp.Predictions[0].GetStructValue().AsMap()["embeddings"].(map[string]interface{})
if !ok {
err = fmt.Errorf("error extracting embeddings")
return
}
values, ok := mapResponse["values"].([]interface{})
if !ok {
err = fmt.Errorf("error extracting embeddings")
return
}
rawEmbeddings := make([]float32, len(values))
for i, v := range values {
rawEmbeddings[i] = float32(v.(float64))
}
embeddings = pgvector.NewVector(rawEmbeddings)
return
}
It should be pointed out that we are not taking into account the model’s input length limitations because we suppose that the report text and the model input are always below 3072 tokens. Anyway, with the autoTruncate parameter set to false, this method will fail if the input length exceeds the limit.
This function can now be used by the final users (for embedding their questions) and by the report generation method, that will create the type.Report (that will be inserted inside the database).
Generate the reports
In Go, we can embed files directly inside the binary using the embed package. Embedding a template is the perfect use-case for this module:
import "embed"
var (
//go:embed templates/daily_report.md
dailyReportTemplate string
)
The method GenerateDailyReport will use gemini-pro to fill the template as requested. After filling the template, we’ll invoke the previously defined GenerateEmbeddings method to completely fill the Report structure previously defined.
// GenerateDailyReport generates a daily report for the given user
func (r *Reporter) GenerateDailyReport(data *UserData) (report *types.Report, err error) {
gemini := r.genaiClient.GenerativeModel("gemini-pro")
temperature := ChatTemperature
gemini.Temperature = &temperature
var builder strings.Builder
fmt.Fprintln(&builder, "This is a markdown template you have to fill with the data I will provide you in the next message.")
fmt.Fprintf(&builder, "```\n%s```\n\n", dailyReportTemplate)
fmt.Fprintln(&builder, "You can find the sections to fill highlighted with \"[LLM to ...]\" with instructions on how to fill the section.")
fmt.Fprintln(&builder, "I will send you the data in JSON format in the next message.")
introductionString := builder.String()
chatSession := gemini.StartChat()
chatSession.History = []*genai.Content{
{
Parts: []genai.Part{
genai.Text(introductionString),
},
Role: "user",
},
{
Parts: []genai.Part{
genai.Text("Send me the data in JSON format. I will fill the template you provided using this data"),
},
Role: "model",
},
}
var jsonData []byte
if jsonData, err = json.Marshal(data); err != nil {
return nil, err
}
var response *genai.GenerateContentResponse
if response, err = chatSession.SendMessage(r.ctx, genai.Text(string(jsonData))); err != nil {
return nil, err
}
report = &types.Report{
StartDate: data.Date,
EndDate: data.Date,
ReportType: "daily",
UserID: r.user.ID,
}
for _, candidates := range response.Candidates {
for _, part := range candidates.Content.Parts {
report.Report += fmt.Sprintf("%s\n", part)
}
}
if report.Embedding, err = r.GenerateEmbeddings(report.Report); err != nil {
return nil, err
}
return report, nil
}
We created a ChatSession with Gemini giving the model a fake history as context and sending the JSON-serialized user data as it’s only source of information.
For example, a (partial) report generated is:
### April 4, 2024
## Activity
- Total Active Time: 41 minutes
- Calories Burned: 346
- Steps Taken: 704
- Distance Traveled: 0 miles
- List of activities:
- Weights: 41 minutes, 346 calories
### Active Minutes Breakdown
- Lightly Active Minutes: 254
- Fairly Active Minutes: 18
- Very Active Minutes: 35
### Heart Rate Zones
- Out of Range: 6 minutes
- Fat Burn: 35 minutes
- Cardio: 0 minutes
- Peak: 0 minutes
## Sleep
- Total Sleep Duration: 7 hours 30 minutes
- Sleep Quality: 75%
- Deep Sleep: 95 minutes
- Light Sleep: 250 minutes
- REM Sleep: 118 minutes
- Time to Fall Asleep: 10 minutes
## Exercise Activities
- Weights: 41 minutes, 346 calories
...
Some information is true, but other information is missing although present in the data (e.g. Cardio/Peak info is present in the JSON but the model inserted 0 as value - that’s wrong). Using an LLM for filling the template is just a way to speed up the template completion process, although being this data available in a structured format the best thing to do would have been to just create the right query to tell the right story. Avoiding thus the randomness of the LLM.
Chatting with the data
Supposing that we have inserted all the reports inside the database, we can now receive messages from the user and try to answer.
Let’s suppose that msg contains the user question. We have to:
- Generate the embeddings
- Search for the best similar reports available (top-k with k=3 just for limiting the context size)
- Share the reports with Gemini inside a chatSession and ask the user question
- Send the result
// 1. Generate the embeddings
var queryEmbeddings pgvector.Vector
if queryEmbeddings, err = reporter.GenerateEmbeddings(msg); err != nil {
return err
}
// 2. Search for similar reports
var reports []string
// Top-3 related reports, sorted by l2 similarity
if err = _db.Model(&types.Report{}).Where(&types.Report{UserID: user.ID})
.Order(fmt.Sprintf("embedding <-> '%s'", queryEmbeddings.String()))
.Select("report").Limit(3).Scan(&reports); err != nil {
return err
}
// 3. Share the reports with Gemini inside a chatSession and ask the user question
builder.Reset() // builder is a stringBuilder
fmt.Fprintln(&builder, "Here are the reports to help you with the analysis:")
fmt.Fprintln(&builder, "")
for _, report := range reports {
fmt.Fprintln(&builder, report)
}
fmt.Fprintln(&builder, "")
fmt.Fprintln(&builder, "Here's the user question you have to answer:")
fmt.Fprintln(&builder, msg)
var responseIterator *genai.GenerateContentResponseIterator = chatSession.SendMessageStream(ctx, genai.Text(builder.String()))
// 4. Send the result
for {
response, err := responseIterator.Next()
if err == iterator.Done {
break
}
for _, candidates := range response.Candidates {
for _, part := range candidates.Content.Parts {
reply := fmt.Sprintf("%s", part)
fmt.Println(reply)
}
}
}
Note that point 3 is partial: we are inside a chatSession where the initial prompt instructed Gemini to behave in a certain way, and that we’ll send messages with reports and the user question.
Point 4 instead is a demonstration of how to receive a streaming response from Gemini - useful when creating a websocket-based application where the Gemini response can be streamed back to the user directly through the websocket.
The image below shows how this interaction allows the users to get insights from their data :)
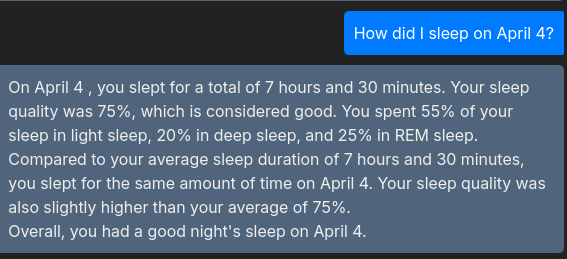
Conclusion & FitSleepInsights
Interacting with Gemini and other models via Vertex AI is quite simple, once understood the pattern to follow and how to extract/insert data from the Protobuf structures. The presented solution that allows the creation of a RAG for data stored in PostgreSQL passes through the generation of a template. This template has been filled by Gemini - but a better solution (although longer to develop) would be to manually fill the template and create these “stories”. In this way, we can remove the randomness of the LLM at least from the data generation part.
The integration of pgvector allowed us to store embeddings on PostgreSQL and make spatial queries in a seamless way.
By the end of the article, we also leaked a screenshot of this feature implemented on fitsleepinsights.app. By the time of the publication of this article, the application is not yet deployed - but the source code is available on Github @ https://github.com/galeone/fitsleepinsights/.
For any feedback or comments, please use the Disqus form below - Thanks!
This article has been possible thanks to the Google ML Developer Programs team that supported this work by providing Google Cloud Credit.